This page was generated from a Jupyter notebook. You can view it on GitHub or download and run it locally.
Normal Inverse Gaussian Distribution
This notebook demonstrates the Normal Inverse Gaussian (NIG) distribution, a normal variance-mean mixture where the mixing distribution is Inverse Gaussian.
Mathematical Definition
The Normal Inverse Gaussian distribution is defined as a normal mixture:
where:
\(\mu \in \mathbb{R}^d\): location parameter
\(\gamma \in \mathbb{R}^d\): skewness parameter
\(\Sigma \in \mathbb{R}^{d \times d}\): covariance scale matrix (positive definite)
\(\delta > 0\): Inverse Gaussian mean parameter (E[Y] = δ)
\(\eta > 0\): Inverse Gaussian shape parameter
Key Properties
Property |
Formula |
|---|---|
Mean |
\(E[X] = \mu + \gamma \delta\) |
Covariance |
\(\text{Cov}[X] = \delta \Sigma + \frac{\delta^3}{\eta} \gamma \gamma^T\) |
Distribution Types
Joint distribution \(f(x, y)\): Exponential family (tractable)
Marginal distribution \(f(x)\): NOT exponential family (requires EM for fitting)
Special Case
NIG is a special case of the Generalized Hyperbolic distribution with GIG parameter \(p = -1/2\).
[1]:
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.gridspec import GridSpec
from scipy import stats
from normix.distributions.mixtures import JointNormalInverseGaussian, NormalInverseGaussian
from normix.distributions.univariate import InverseGaussian
from normix.utils import (
plot_joint_distribution_1d,
plot_marginal_distribution_2d,
validate_moments,
print_moment_validation,
fit_and_track_convergence,
plot_em_convergence,
test_joint_fitting,
print_fitting_results
)
plt.style.use('seaborn-v0_8-whitegrid')
%matplotlib inline
# Consistent styling
COLORS = {'primary': 'tab:blue', 'secondary': 'tab:red', 'tertiary': 'tab:green'}
np.set_printoptions(precision=4, suppress=True)
Parameter Sets for Testing
We test with three different parameter configurations to demonstrate the distribution’s flexibility.
[2]:
# Define three parameter sets for comprehensive testing
PARAM_SETS = [
{
'name': 'Symmetric (γ=0)',
'mu': np.array([0.0, 0.0]),
'gamma': np.array([0.0, 0.0]),
'sigma': np.array([[1.0, 0.3], [0.3, 1.0]]),
'delta': 1.0,
'eta': 1.0
},
{
'name': 'Right-skewed (γ>0)',
'mu': np.array([0.0, 0.0]),
'gamma': np.array([0.5, 0.3]),
'sigma': np.array([[1.0, 0.2], [0.2, 1.5]]),
'delta': 1.5,
'eta': 2.0
},
{
'name': 'Left-skewed (γ<0)',
'mu': np.array([1.0, -0.5]),
'gamma': np.array([-0.4, 0.2]),
'sigma': np.array([[2.0, -0.5], [-0.5, 1.0]]),
'delta': 2.0,
'eta': 1.5
}
]
# Display parameter sets
for i, params in enumerate(PARAM_SETS):
print(f"\nParameter Set {i+1}: {params['name']}")
print(f" μ = {params['mu']}")
print(f" γ = {params['gamma']}")
print(f" δ = {params['delta']}, η = {params['eta']}")
Parameter Set 1: Symmetric (γ=0)
μ = [0. 0.]
γ = [0. 0.]
δ = 1.0, η = 1.0
Parameter Set 2: Right-skewed (γ>0)
μ = [0. 0.]
γ = [0.5 0.3]
δ = 1.5, η = 2.0
Parameter Set 3: Left-skewed (γ<0)
μ = [ 1. -0.5]
γ = [-0.4 0.2]
δ = 2.0, η = 1.5
Part 1: Joint Distribution (1D X)
The joint distribution \(f(x, y)\) is an exponential family with natural parameters.
[3]:
def get_1d_params(params):
"""Extract 1D version of parameters."""
return {
'mu': np.array([params['mu'][0]]),
'gamma': np.array([params['gamma'][0]]),
'sigma': np.array([[params['sigma'][0, 0]]]),
'delta': params['delta'],
'eta': params['eta']
}
1.1 Joint Distribution Visualization
[4]:
N_SAMPLES = 5000
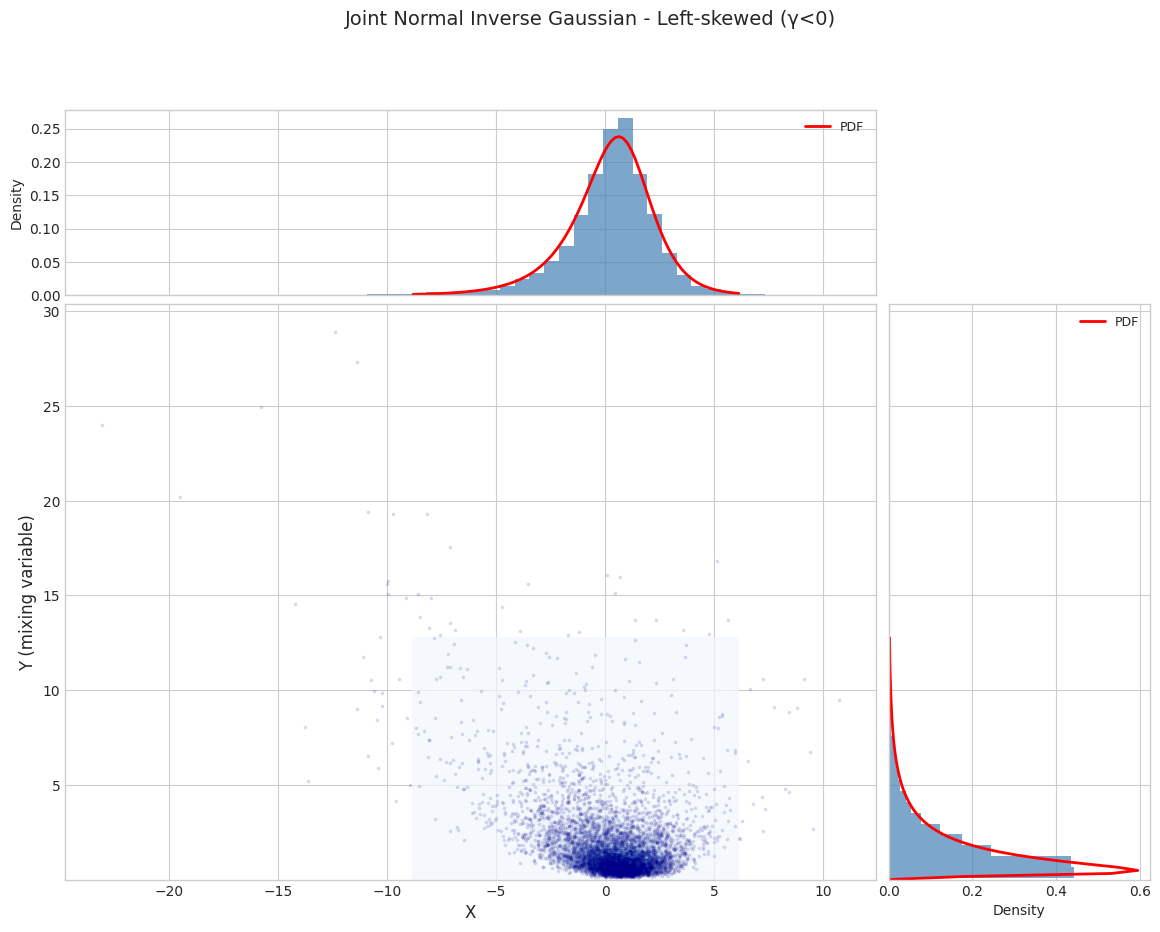
for i, params in enumerate(PARAM_SETS):
params_1d = get_1d_params(params)
joint_dist = JointNormalInverseGaussian.from_classical_params(**params_1d)
print(f"\n{'='*70}")
print(f"Parameter Set {i+1}: {params['name']}")
print(f"{'='*70}")
fig = plot_joint_distribution_1d(
joint_dist,
n_samples=N_SAMPLES,
random_state=42,
title=f"Joint Normal Inverse Gaussian - {params['name']}"
)
plt.show()
======================================================================
Parameter Set 1: Symmetric (γ=0)
======================================================================
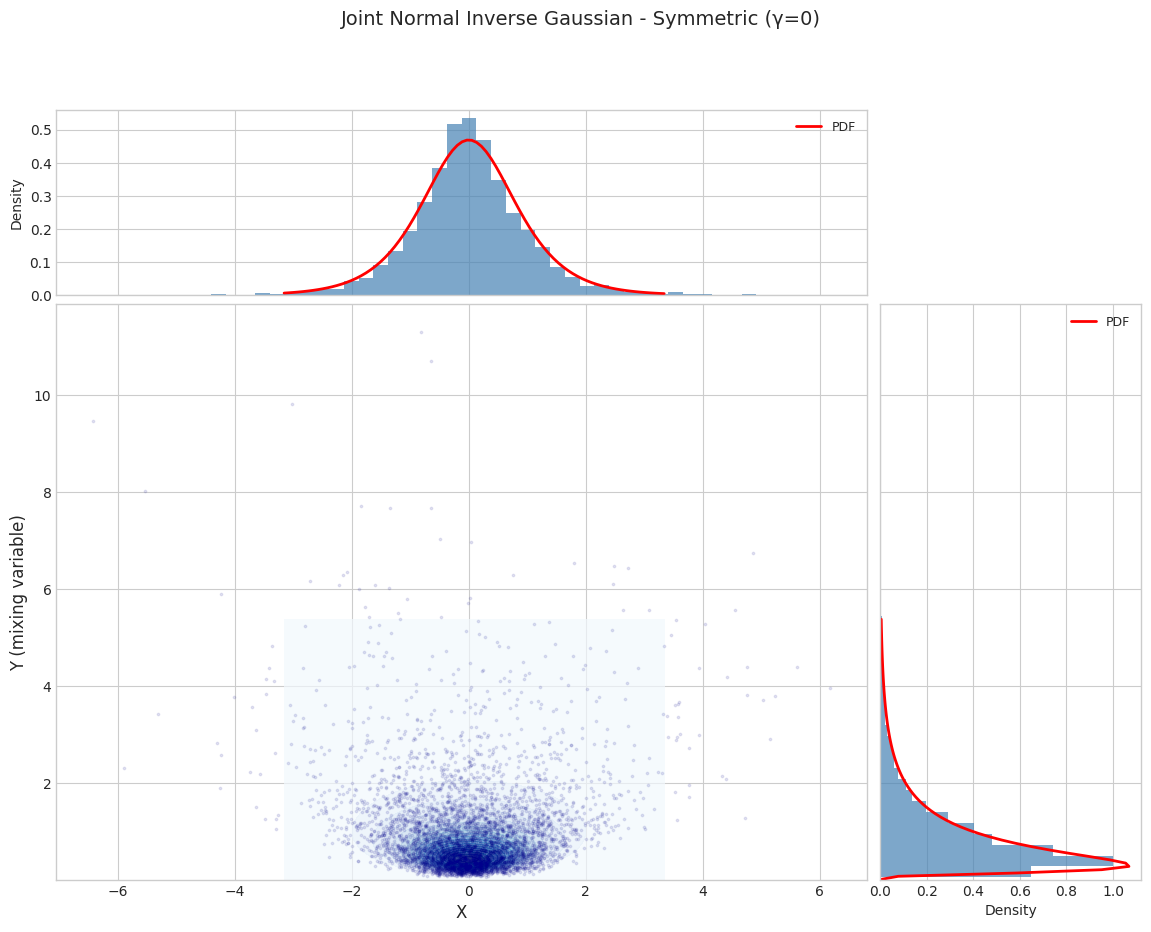
======================================================================
Parameter Set 2: Right-skewed (γ>0)
======================================================================
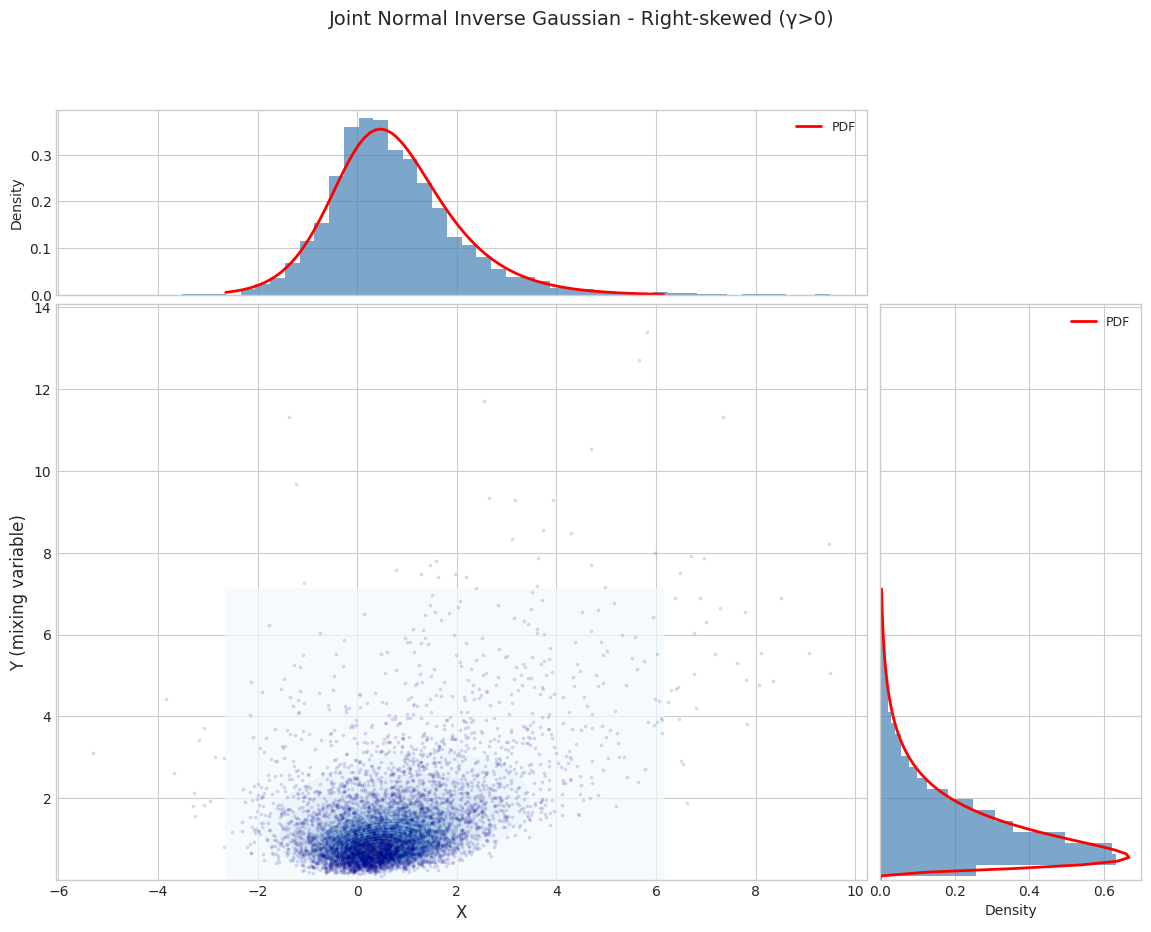
======================================================================
Parameter Set 3: Left-skewed (γ<0)
======================================================================
1.2 Moment Validation (Joint)
[5]:
for i, params in enumerate(PARAM_SETS):
params_1d = get_1d_params(params)
joint_dist = JointNormalInverseGaussian.from_classical_params(**params_1d)
results = validate_moments(joint_dist, n_samples=50000, random_state=42, is_joint=True)
print_moment_validation(results, f"Joint NIG - {params['name']}")
============================================================
Moment Validation: Joint NIG - Symmetric (γ=0)
============================================================
X_mean : sample = -0.0009, theory = 0.0000, rel_err = 9.03e+06
Y_mean : sample = 1.0028, theory = 1.0000, rel_err = 2.79e-03
X_var : sample = 1.0206, theory = 1.0000, rel_err = 2.06e-02
Y_var : sample = 1.0209, theory = 1.0000, rel_err = 2.09e-02
============================================================
Moment Validation: Joint NIG - Right-skewed (γ>0)
============================================================
X_mean : sample = 0.7513, theory = 0.7500, rel_err = 1.68e-03
Y_mean : sample = 1.5044, theory = 1.5000, rel_err = 2.91e-03
X_var : sample = 1.9634, theory = 1.9219, rel_err = 2.16e-02
Y_var : sample = 1.7077, theory = 1.6875, rel_err = 1.20e-02
============================================================
Moment Validation: Joint NIG - Left-skewed (γ<0)
============================================================
X_mean : sample = 0.1963, theory = 0.2000, rel_err = 1.83e-02
Y_mean : sample = 2.0050, theory = 2.0000, rel_err = 2.49e-03
X_var : sample = 4.9320, theory = 4.8533, rel_err = 1.62e-02
Y_var : sample = 5.4041, theory = 5.3333, rel_err = 1.33e-02
1.3 Joint Distribution Fitting (Exponential Family MLE)
[6]:
for i, params in enumerate(PARAM_SETS):
params_1d = get_1d_params(params)
fitted_dist, fitted_params, param_errors = test_joint_fitting(
JointNormalInverseGaussian,
params_1d,
n_samples=N_SAMPLES,
random_state=42
)
print_fitting_results(params_1d, fitted_params, param_errors, f"Joint NIG - {params['name']}")
============================================================
Fitting Results: Joint NIG - Symmetric (γ=0)
============================================================
Parameter True Fitted Rel.Error
------------------------------------------------------------
mu 0.0000 -0.0141 1.41e+08
gamma 0.0000 0.0137 1.37e+08
sigma 1.0000 0.9988 1.20e-03
delta 1.0000 0.9806 1.94e-02
eta 1.0000 1.0179 1.79e-02
============================================================
Fitting Results: Joint NIG - Right-skewed (γ>0)
============================================================
Parameter True Fitted Rel.Error
------------------------------------------------------------
mu 0.0000 -0.0177 1.77e+08
gamma 0.5000 0.5106 2.12e-02
sigma 1.0000 0.9988 1.16e-03
delta 1.5000 1.4772 1.52e-02
eta 2.0000 2.0356 1.78e-02
============================================================
Fitting Results: Joint NIG - Left-skewed (γ<0)
============================================================
Parameter True Fitted Rel.Error
------------------------------------------------------------
mu 1.0000 0.9762 2.38e-02
gamma -0.4000 -0.3884 2.91e-02
sigma 2.0000 1.9976 1.19e-03
delta 2.0000 1.9628 1.86e-02
eta 1.5000 1.5267 1.78e-02
Part 2: Marginal Distribution (2D X)
The marginal distribution \(f(x) = \int f(x, y) dy\) is NOT an exponential family.
Fitting requires the EM algorithm.
2.1 Marginal Distribution Visualization
[7]:
for i, params in enumerate(PARAM_SETS):
marginal_dist = NormalInverseGaussian.from_classical_params(**params)
print(f"\n{'='*70}")
print(f"Parameter Set {i+1}: {params['name']}")
print(f"{'='*70}")
fig = plot_marginal_distribution_2d(
marginal_dist,
n_samples=N_SAMPLES,
random_state=42,
title=f"Marginal Normal Inverse Gaussian (2D) - {params['name']}"
)
plt.show()
======================================================================
Parameter Set 1: Symmetric (γ=0)
======================================================================
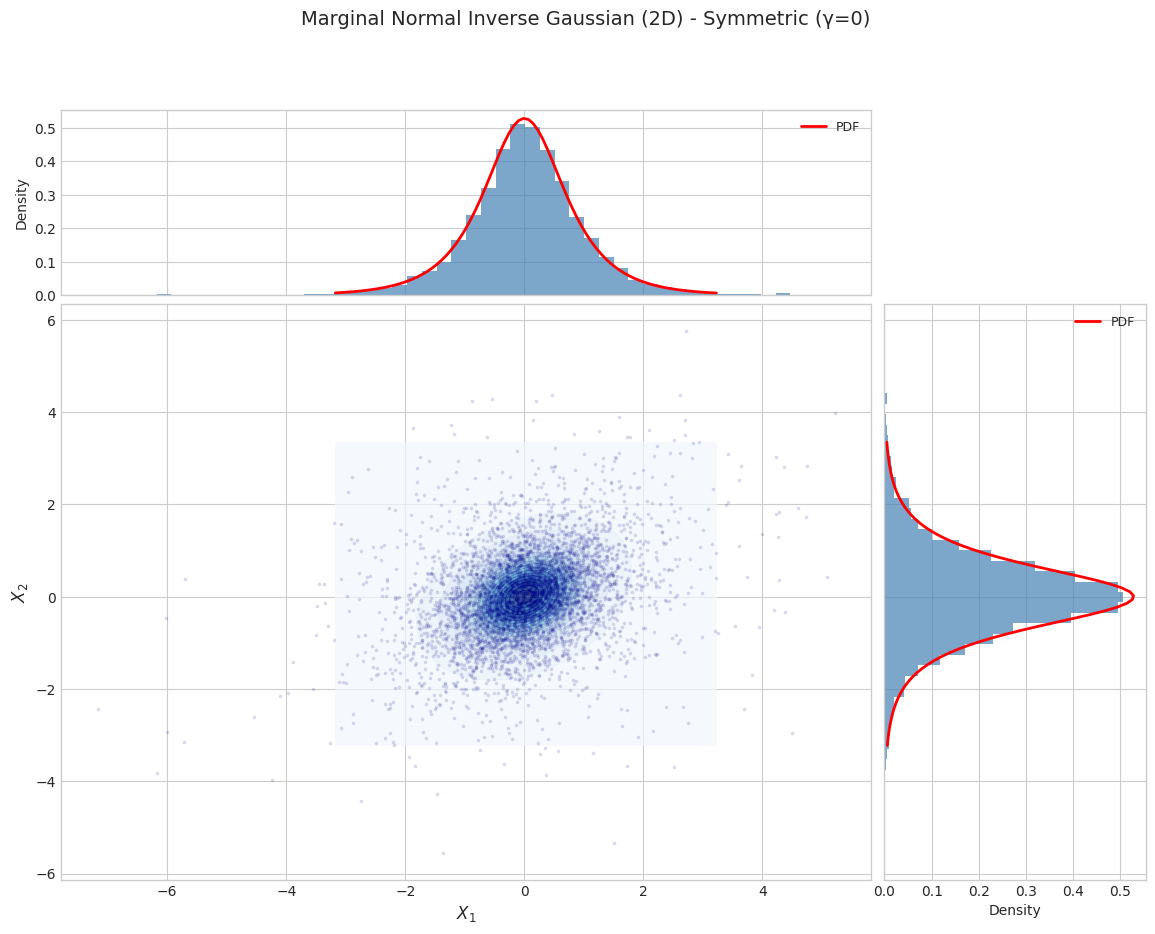
======================================================================
Parameter Set 2: Right-skewed (γ>0)
======================================================================
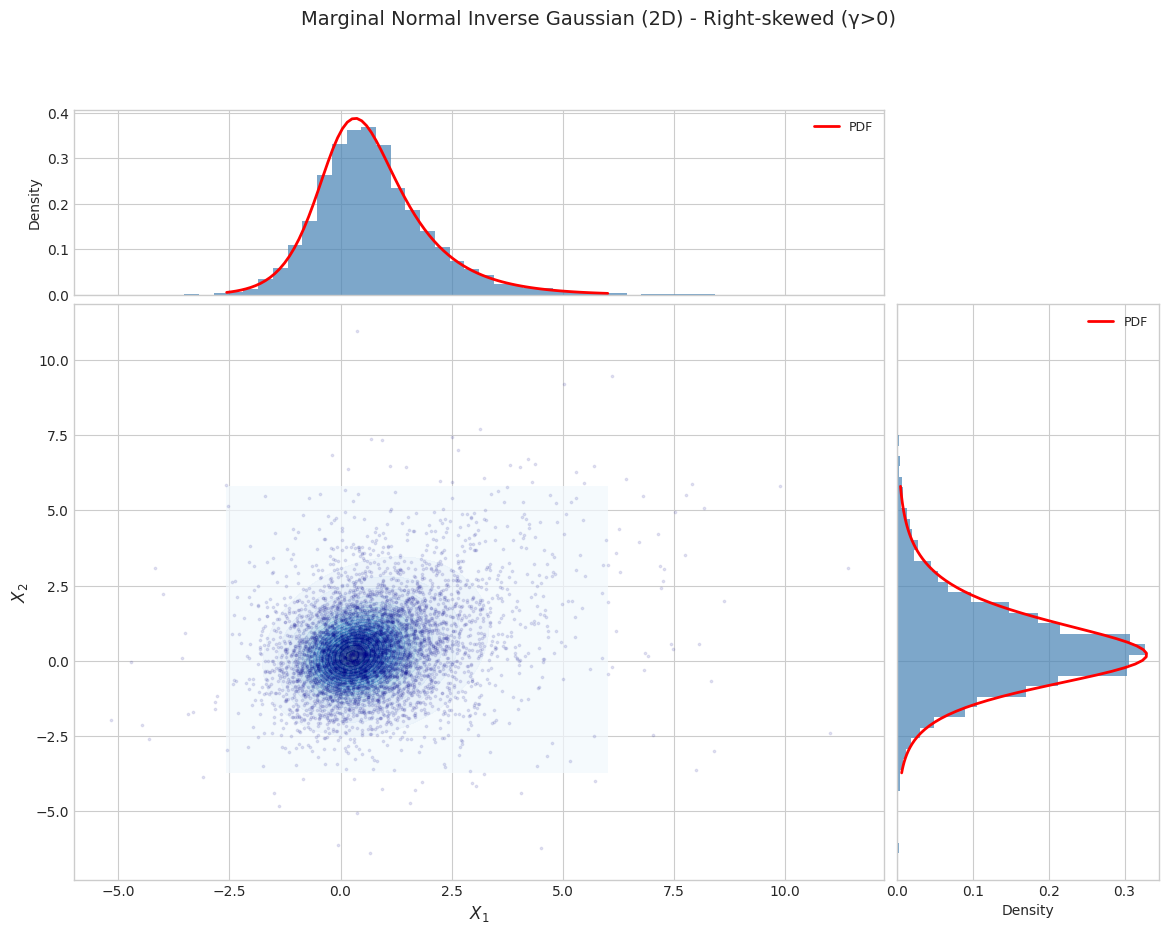
======================================================================
Parameter Set 3: Left-skewed (γ<0)
======================================================================
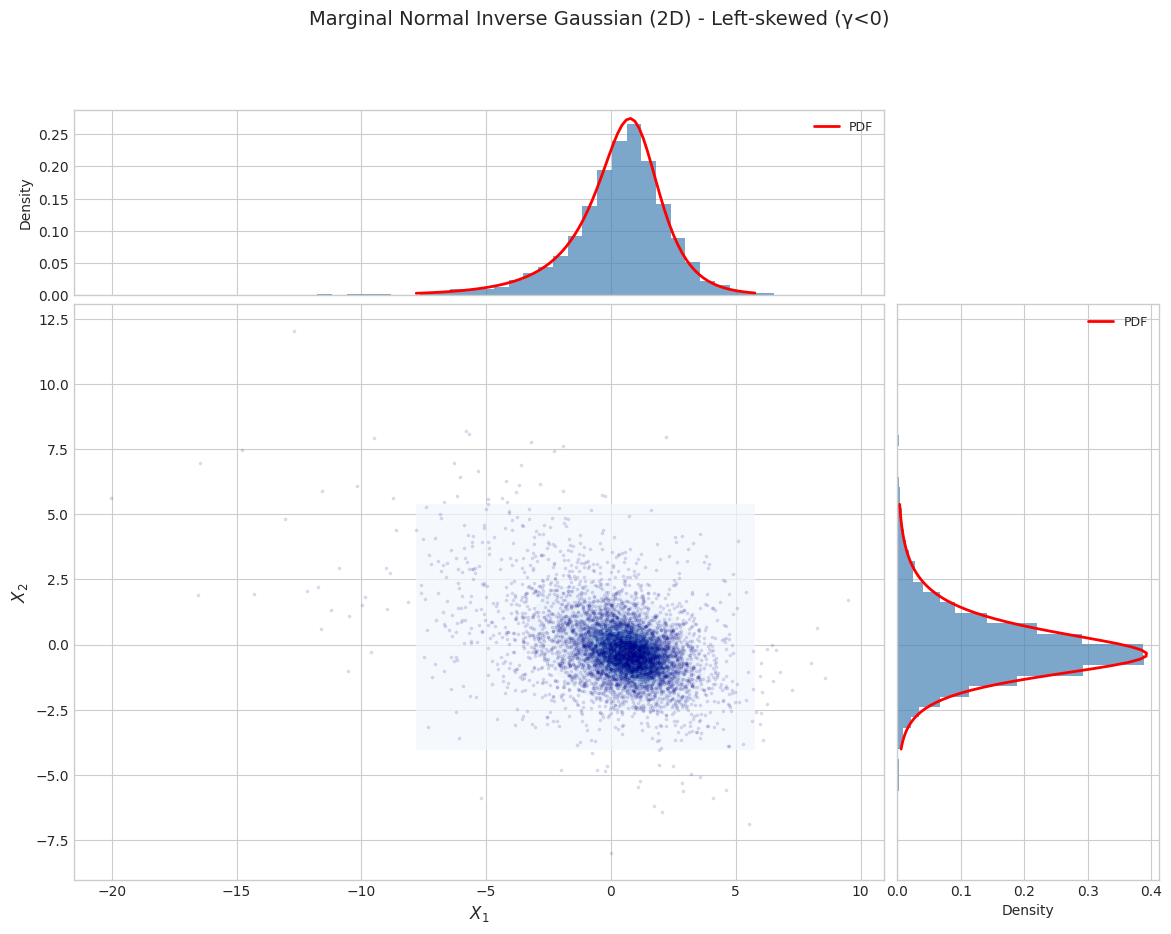
2.2 Moment Validation (Marginal)
[8]:
for i, params in enumerate(PARAM_SETS):
marginal_dist = NormalInverseGaussian.from_classical_params(**params)
results = validate_moments(marginal_dist, n_samples=50000, random_state=42, is_joint=False)
print_moment_validation(results, f"Marginal NIG - {params['name']}")
============================================================
Moment Validation: Marginal NIG - Symmetric (γ=0)
============================================================
mean :
sample = [0.0048 0.0031]
theory = [0. 0.]
rel_err = [47529792.35 31132175.93]
variance :
sample = [1.0095 1.003 ]
theory = [1. 1.]
rel_err = [0.01 0. ]
============================================================
Moment Validation: Marginal NIG - Right-skewed (γ>0)
============================================================
mean :
sample = [0.7577 0.4542]
theory = [0.75 0.45]
rel_err = [0.01 0.01]
variance :
sample = [1.9515 2.4097]
theory = [1.9219 2.4019]
rel_err = [0.02 0. ]
============================================================
Moment Validation: Marginal NIG - Left-skewed (γ<0)
============================================================
mean :
sample = [ 0.2067 -0.0984]
theory = [ 0.2 -0.1]
rel_err = [0.03 0.02]
variance :
sample = [4.8635 2.2215]
theory = [4.8533 2.2133]
rel_err = [0. 0.]
2.3 EM Algorithm Fitting with Convergence Tracking
[9]:
for i, params in enumerate(PARAM_SETS):
print(f"\n{'='*70}")
print(f"Parameter Set {i+1}: {params['name']}")
print(f"{'='*70}")
# Generate data from true distribution
true_dist = NormalInverseGaussian.from_classical_params(**params)
X_data = true_dist.rvs(size=N_SAMPLES, random_state=42)
# Fit with EM and track convergence
fitted_dist, convergence = fit_and_track_convergence(
NormalInverseGaussian,
X_data,
max_iter=100,
random_state=43
)
# Print results
print(f"\nConverged: {convergence.converged}")
print(f"Iterations: {len(convergence.iterations)}")
if convergence.log_likelihoods:
print(f"Initial LL: {convergence.log_likelihoods[0]:.4f}")
print(f"Final LL: {convergence.log_likelihoods[-1]:.4f}")
# Compare parameters
print("\nParameter Comparison:")
print(f" True δ = {params['delta']:.4f}, Fitted δ = {convergence.final_params['delta']:.4f}")
print(f" True η = {params['eta']:.4f}, Fitted η = {convergence.final_params['eta']:.4f}")
# Plot convergence
if convergence.iterations:
fig = plot_em_convergence(
convergence,
title=f"EM Convergence - {params['name']}"
)
plt.show()
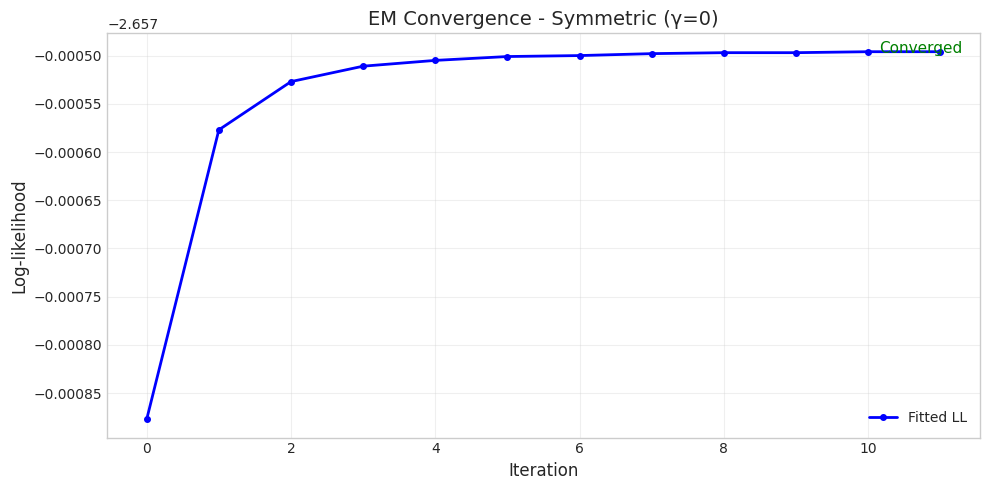
======================================================================
Parameter Set 1: Symmetric (γ=0)
======================================================================
Converged: True
Iterations: 12
Initial LL: -2.6579
Final LL: -2.6575
Parameter Comparison:
True δ = 1.0000, Fitted δ = 1.0040
True η = 1.0000, Fitted η = 0.9793
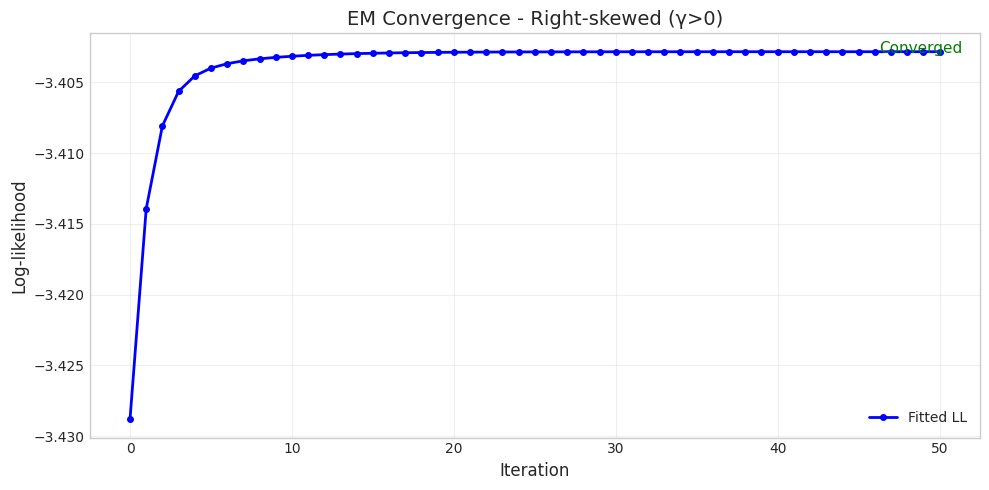
======================================================================
Parameter Set 2: Right-skewed (γ>0)
======================================================================
Converged: True
Iterations: 51
Initial LL: -3.4288
Final LL: -3.4028
Parameter Comparison:
True δ = 1.5000, Fitted δ = 0.9257
True η = 2.0000, Fitted η = 1.1585
======================================================================
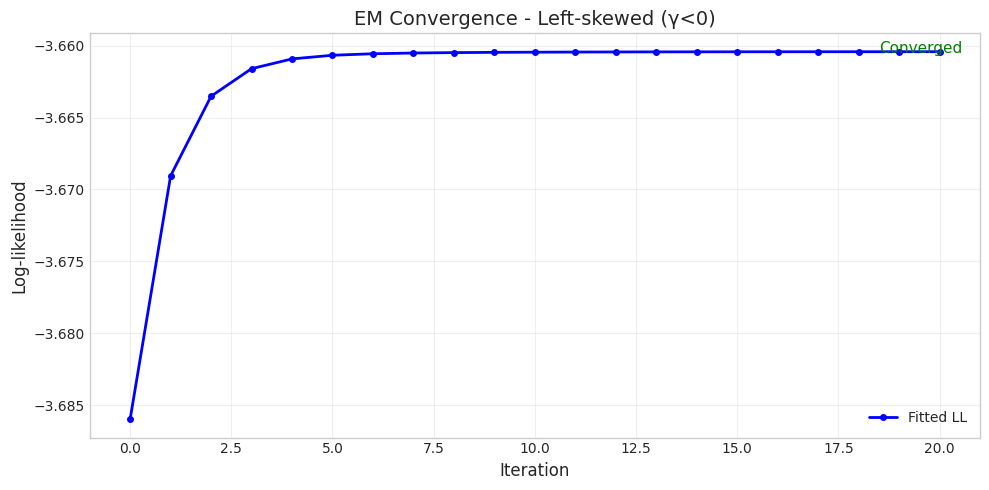
Parameter Set 3: Left-skewed (γ<0)
======================================================================
Converged: True
Iterations: 21
Initial LL: -3.6860
Final LL: -3.6604
Parameter Comparison:
True δ = 2.0000, Fitted δ = 1.0162
True η = 1.5000, Fitted η = 0.7952
Part 3: Exponential Family Structure
The joint distribution has the exponential family form:
[11]:
# Demonstrate exponential family structure
params_1d = get_1d_params(PARAM_SETS[1]) # Use right-skewed set
joint_dist = JointNormalInverseGaussian.from_classical_params(**params_1d)
print("Exponential Family Structure")
print("="*60)
print(f"\nClassical parameters:")
print(joint_dist.classical_params)
print(f"\nNatural parameters θ:")
print(joint_dist.natural_params)
print(f"\nExpectation parameters η = E[t(X,Y)]:")
print(joint_dist.expectation_params)
Exponential Family Structure
============================================================
Classical parameters:
{'mu': array([0.]), 'gamma': array([0.5]), 'sigma': array([[1.]]), 'delta': np.float64(1.5), 'eta': np.float64(2.0)}
Natural parameters θ:
[-2. -2. -1.0139 0.5 0. -0.5 ]
Expectation parameters η = E[t(X,Y)]:
[0.1172 1.1667 1.5 0.75 0.5 1.375 ]
[13]:
# Verify E[t(X,Y)] matches expectation parameters
X_samples, Y_samples = joint_dist.rvs(size=50000, random_state=42)
t_samples = joint_dist._sufficient_statistics(X_samples, Y_samples)
eta_sample = np.mean(t_samples, axis=0)
eta_theory = joint_dist.expectation_params
print("\nVerification: E[t(X,Y)] from samples vs theory")
print("="*60)
print(f"Sample: {eta_sample}")
print(f"Theory: {eta_theory}")
print(f"Max error: {np.max(np.abs(eta_sample - eta_theory)):.6f}")
Verification: E[t(X,Y)] from samples vs theory
============================================================
Sample: [0.1178 1.1691 1.5044 0.7513 0.4983 1.3865]
Theory: [0.1172 1.1667 1.5 0.75 0.5 1.375 ]
Max error: 0.011455
Summary
The Normal Inverse Gaussian distribution is a flexible model with:
Feature |
Description |
|---|---|
Mixing |
\(Y \sim \text{InvGauss}(\delta, \eta)\) |
Skewness |
Controlled by \(\gamma\) |
Tail behavior |
Semi-heavy tails |
Joint fitting |
Exponential family MLE |
Marginal fitting |
EM algorithm |
Special case |
GH with \(p = -1/2\) |